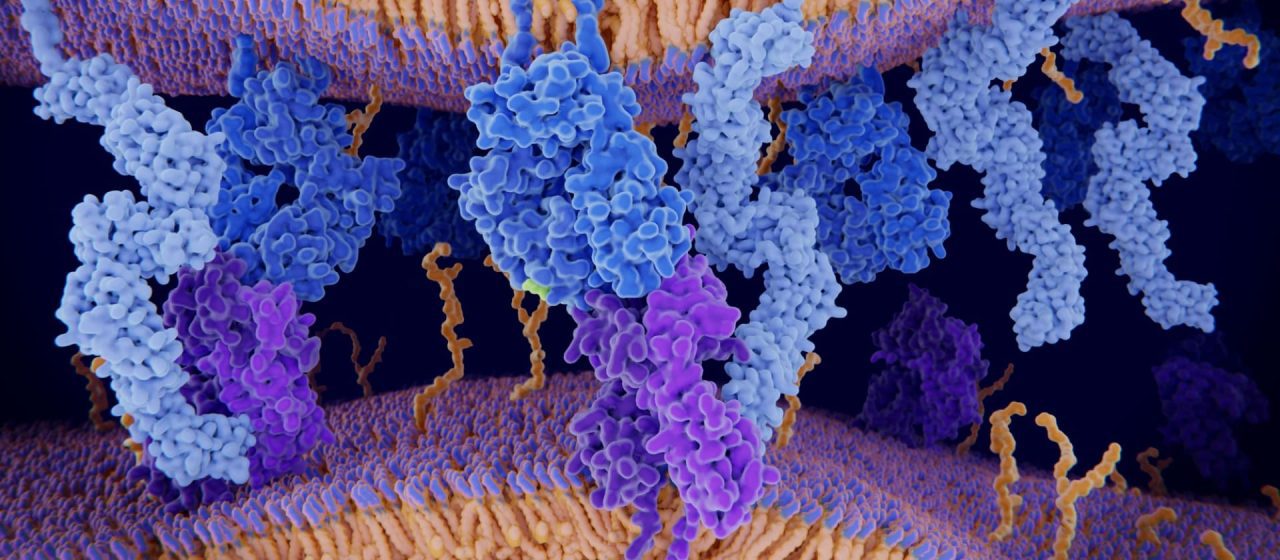
Professor Giri Narasimhan and Dr. Vitalii Stebliankin created a machine-learning model to predict how protein molecules will bind together, important for designing new medications and vaccines. The AI-based method uses biological and structural information to score the strength of the bond, giving scientists important starting information to figure out how to build a medication to bind to a specific protein. The results were recently published in Nature Machine Intelligence, and are featured in a news article at FIU.
READ MORE: https://news.fiu.edu/2023/matchmaking-with-ai-to-help-proteins-pair-up
